Plan Overview: This project will inform critical knowledge gaps on how river diversions influence the ecological trajectory and functionality of natural and created marshes by assessing species composition and abundances, and characterizing aquatic food web structure and fish residency, across salinity gradients in the West Pointe à la Hache (WPH) siphon area. This improved understanding of trophic relationships will inform and parameterize an ecosystem model to aid in predicting relationships between habitat, salinity and community composition and food web structure in natural and created marshes.
The goal of this research is to guide future restoration effort by integrating community and food-web approaches into management and restoration planning.
Main Project Objectives

Examine species composition, relative abundances, and food web structure in natural marshes along a salinity gradient influenced by a river diversion.

Examine species composition, relative abundances, and food web structure across different-aged created marshes that are influenced by a river diversion.
Use the field data gathered above to build and test a food web model (Ecopath with Ecosim) that can be used to predict the impact of habitat restoration on marsh food webs.
Research Approaches
Species Abundances and Community Structure:

Primary Producers and Nutrient Concentrations: Marsh platform primary producer biomass and diversity as well as benthic microalgal biomass are quantified along with phytoplankton biomass in the channels adjacent to the marsh. Additionally, marsh soil and adjacent channel water are characterized in terms of salinity, temperature, pH, and carbon and nutrient concentrations.
Microbiology: Soil cores, sediment cores, and water samples are collected for analysis of relative and quantitative abundances for bacterial, archaea, and fungi (referred to as total microbial diversity), total environmental DNA, and microbial biomass. Data describe changes in microbial diversity, relative abundances, and community structure that may be similar or different among the sampled natural and created marshes.
Infaunal benthos: Infaunal benthos (> 0.5 mm) will be collected using core samples, preserved, and identified to lowest possible taxon. From these fundamental data, there will be a determination of mean species richness, abundance, diversity, and estimates of biomass for food web structure.
Macroinvertebrates: Epibenthic invertebrates (crabs, bivalves, and gastropods) will be collected, near the core samples and at the marsh edge. Specifically, density analysis will focus on three representative intertidal macroinvertebrates, Fiddler crabs, marsh periwinkles, and Gulf ribbed mussels. Crab burrow density is used as a proxy for crab abundance with burrow diameter as a proxy for crab size.
Aquatic and terrestrial insects and spiders: Terrestrial arthropods (insects and spiders) are sampled on plots using sweep nets. Emergence traps and aquatic net sampling are also incorporated to capture aquatic insects at the marsh edge and platform. Samples are frozen in the field and identified in the laboratory to the species level.
Fish: Marsh fish and nekton composition and abundance (catch per unit effort) are quantified using fyke nets and wire mesh traps. Off-marsh fish and nekton communities will be quantitatively sampled using replicated tows of a 5 m otter trawl behind a small research vessel. All captured specimens are identified to the lowest taxonomic level possible, measured, and subsampled for isotopic content as needed. Otoliths will be extracted from some samples.
Fish Residency:
Otolith chemistry is used to reconstruct the salinity regime experienced by marsh-resident fish and off-marsh juvenile fish over their lifetimes. An extracted left- or right-side otolith is used for laser ablation and mass spectrometery to generate a geochemical time series experienced by the fish through its lifetime. The other otolith is used to compare fish growth between natural vs. restored marshes and is estimated using an analysis of daily growth rings, analysis of annual otolith width increments, or use of magnesium (Mg) as a proxy for fish growth.
Stable Isotope Analysis:

A range of marsh primary producers, and invertebrate and vertebrate consumers are sampled from the marshes (see Species Abundances and Community Structure, above). Bulk tissue carbon (δ13C), nitrogen (δ15N), and sulfur (δ34S) isotopes, and amino acid-specific stable isotopes (δ13C) help identify differences in food-web structure and carbon flow and variation among seasons and relative distance from the WPH siphon.
Primary producers and infauna will be analyzed whole, only muscle tissue will be analyzed for all other consumers. Bulk isotopic samples are analyzed through an interfaced Thermo Delta V Advantage CFIRMS, and δ13C CSIA-AA values are obtained via Gas Chromatography-IRMS following acid hydrolysis and derivatization.
Bayesian stable isotope mixing models are applied to estimate the percent contribution of organic matter sources by species, and IsoWeb is used to compare and contrast food web topologies based on bulk isotope data. Consumer trophic levels are calculated using static and scaled trophic enrichment factors and estimates of basal food web sourcesThis approach generates isotopic niche areas and volumes for each species in each food web. Changes in the size, overlap, and position of species and community level niche areas in isotopic space and changes in overall food chain length and carbon source usage are used to infer marsh food web structure and function along salinity gradients and among different-aged created marshes under the influences of the WPH siphon. Amino acid-specific (δ13C) values and Bayesian stable isotope mixing models are used to estimate the percent contribution of basal carbon sources that support a subset of marsh consumers (e.g., penaeid shrimp, periwinkle snails, gulf killifish, and blue crabs). These data identify changes in carbon sources along a salinity gradient and over time in created marshes.
Integration and Food Web Modeling:
Interactions among species make it difficult to predict the results of restoration efforts from habitat models alone. The strength of species interactions plays an important role in conferring ecosystem stability, with weak trophic interactions tending to increase ecosystem stability and resilience. Quantifying differences in predator-prey interactions between natural and restored marshes provides insight on ecosystem resilience.
Data from the field sampling and biomarker studies are used to parameterize two base Ecopath models (one for natural and one for restored marshes) by modifying the diet matrix of an existing model for nearby Breton Sound trophic diversity/redundancy, a critical component of food web resilience.
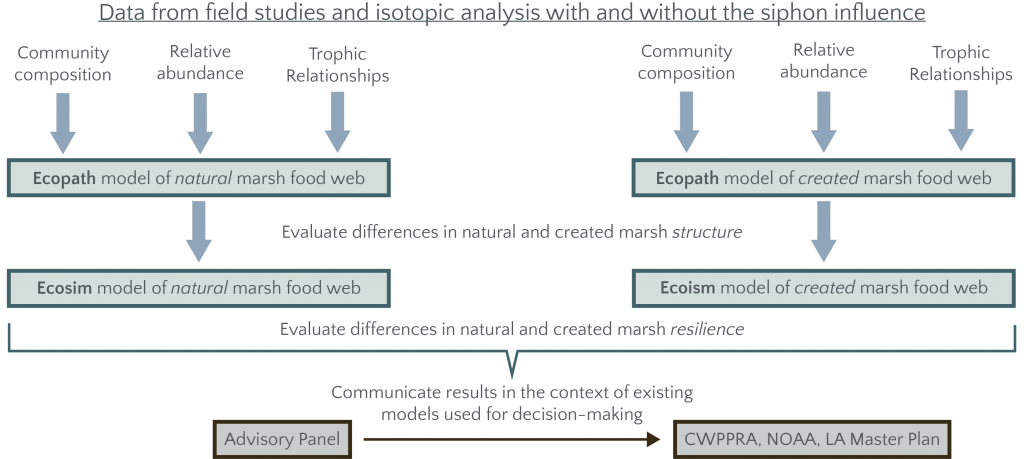
Base Ecopath models are used to identify emergent characteristics for created vs. natural marshes. Proposed metrics are net primary production, system total respiration, total biomass flows and total system throughput. Ecosystem properties of interest are ecosystem maturity, Kempton’s Q metric of biodiversity modified for Ecopath, pelagic:demersal ratio and the evenness of functional group biomass by Shannon’s entropy index. [de Mutsert, K. et al. 2012, Marine and Coastal Fisheries 4: 104–116, https://doi.org/10.1080/19425120.2012.672366]
A dynamic food web model (Ecosim) is used to understand the comparative resilience of natural and restored marsh food webs to different simulated perturbations, including: loss of individual top predator species, loss of species identified as most sensitive to salinity shifts and loss of species identified as critical network nodes (i.e., keystone species) from the Ecopath analysis. Resilience is assessed through percent change in each of the unperturbed components of the model at equilibrium and return time (time to reach 95% of new equilibrium biomass).
Translation to Management
One of the primary goals of this research is to guide future restoration efforts by integrating community and food-web approaches into management and restoration planning.
- Compare with community and species level models commonly used by CWPPRA to evaluate marsh restoration projects.
- Identify the insights mangers can gain from metrics of food web structure that can help interpret and refine current models and practices.
Project Advisory Board Members:
- Stuart Brown, Coastal Protection and Restoration Authority, Baton Rouge, LA
- Jim Pahl, Coastal Protection and Restoration Authority, Baton Rouge, LA
- Sharon Osowski, Marine, Coastal and Analysis Section (6WQ-EC), US EPA Region 6, Dallas, TX
- Kevin Roy, U.S. Fish and Wildlife Service, Louisiana Ecological Services Office, Lafayette, LA
- Robert Spears, Plaquemines Parish Coastal Zone Management Office, Belle Chasse, LA
- Pat Williams, NOAA Restoration Center, Baton Rouge, LA
NOAA RESTORE Program Officers:
- Frank Parker III, Associate Director, NOAA RESTORE Science Program, NOAA/NOS/NCCOS
- Melissa Carle, Monitoring and Planning Coordinator, Deepwater Horizon Restoration, NOAA Restoration Center
- Shannon Martin, Ecosystem Assessment Scientist, Cooperative Institute for Marine and Atmospheric Studies, CIMAS/NOAA/OAR/AOML